Normalization¶
In VRS, “normalization” refers to the process of rewriting an ambiguous variation representation of variation into a canonical form. Normalization eliminates a class of ambiguity that impedes comparison of variation across systems.
In the sequencing community, “normalization” refers to the process of converting a given sequence variant into a canonical form, typically by left- or right-shuffling insertion/deletion variants. VRS normalization extends this concept to all classes of VRS Variation objects.
Implementations MUST provide a normalize function that accepts any Variation object and returns a normalized Variation. Guidelines for these functions are below.
General Normalization Rules¶
Object types that do not have explicit VRS normalization rules below are returned as-is. That is, all types of Variation MUST be supported, even if such objects are unchanged.
VRS normalization functions are idempotent: Normalizing a previously-normalized object returns an equivalent object.
VRS normalization functions are not necessarily homomorphic: That is, the input and output objects may be of different types.
Allele Normalization¶
Certain insertion or deletion alleles may have ambiguous representations when using conventional sequence normalization, resulting in significant challenges when comparing such alleles.
VRS uses a “fully-justified” normalization algorithm adapted from NCBI’s Variant Overprecision Correction Algorithm 1. Fully-justified normalization expands such ambiguous representation over the entire region of ambiguity, resulting in an unambiguous representation that may be readily compared with other alleles.
This algorithm was designed for Allele instances in which the Reference Allele Sequence and Alternate Allele Sequence are precisely known and intended to be normalized. In some instances, this may not be desired, e.g. faithfully maintaining a sequence represented as a repeating subsequence through a RepeatSequenceExpression object. We also anticipate that these edge cases will not be common, and encourage adopters to use the VRS Allele Normalization Algorithm whenever possible.
LiteralSequenceExpression Alleles¶
When normalizing an Allele with a LiteralSequenceExpression state, the following normalization rules apply:
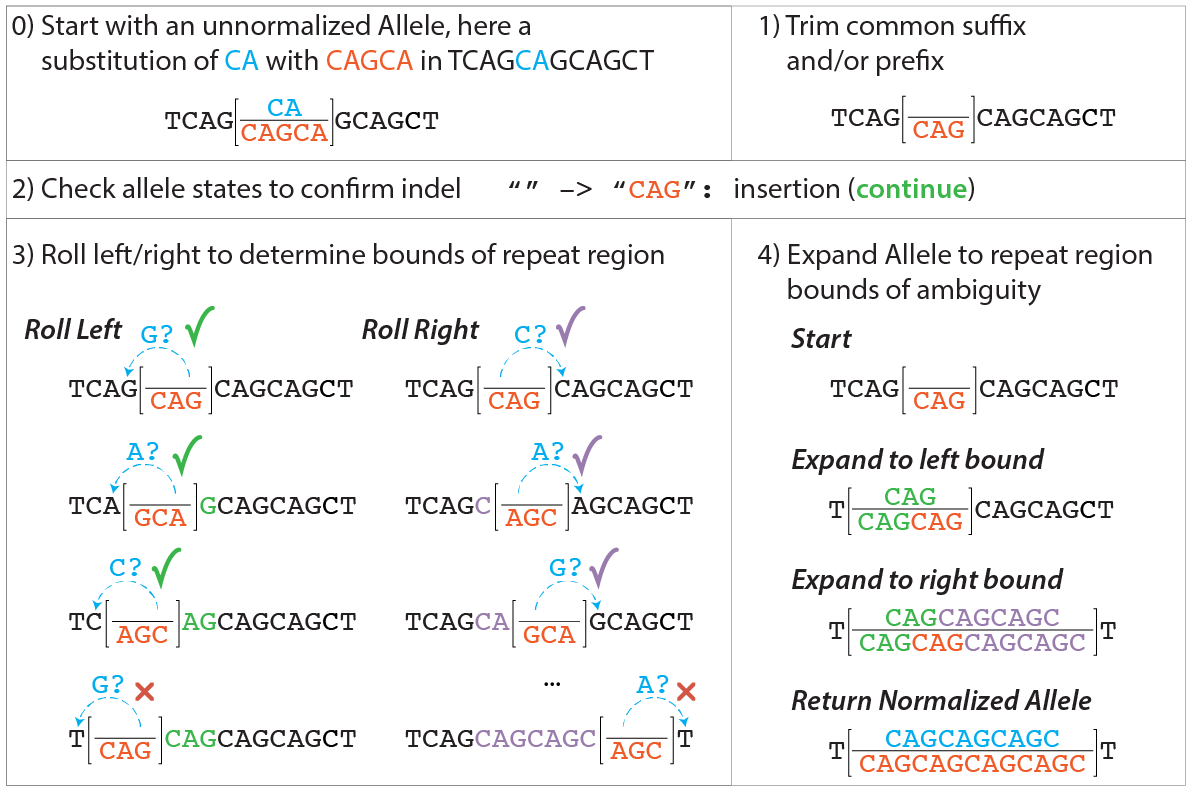
Start with an unnormalized Allele, with corresponding reference and alternate Allele Sequences.
The Reference Allele Sequence refers to the subsequence at the Allele SequenceLocation.
The Alternate Allele Sequence refers to the Sequence described by the Allele state attribute.
Let start and end initially be the start and end of the Allele SequenceLocation.
Trim common flanking sequence from Allele sequences.
Trim common suffix sequence (if any) from both of the Allele Sequences and decrement end by the length of the trimmed suffix.
Trim common prefix sequence (if any) from both of the Allele Sequences and increment start by the length of the trimmed prefix.
Compare the two Allele sequences, if:
both are empty, the input Allele is a reference Allele. Return the input Allele unmodified.
both are non-empty, the input Allele has been normalized to a substitution. Return a new Allele with the modified start, end, and Alternate Allele Sequence.
one is empty, the input Allele is an insertion (empty reference sequence) or a deletion (empty alternate sequence). Continue to step 3.
Determine bounds of ambiguity.
Left roll: Set a left_roll_bound equal to start. While the terminal base of the non-empty Allele sequence is equal to the base preceding the left_roll_bound, decrement left_roll_bound and circularly permute the Allele sequence by removing the last character of the Allele sequence, then prepending the character to the resulting Allele sequence.
Right roll: Set a right_roll_bound equal to start. While the terminal base of the non-empty Allele sequence is equal to the base following the right_roll_bound, increment right_roll_bound and circularly permute the Allele sequence by removing the first character of the Allele sequence, then appending the character to the resulting Allele sequence.
Construct a new Allele covering the entire region of ambiguity.
Prepend characters from left_roll_bound to start to both Allele Sequences.
Append characters from start to right_roll_bound to both Allele Sequences.
Set start to left_roll_bound and end to right_roll_bound, and return a new Allele with the modified start, end, and Alternate Allele Sequence.
RepeatedSequenceExpression Alleles¶
When normalizing an Allele with a RepeatedSequenceExpression state, normalization is similar to that of LiteralSequenceExpression, expanding the Reference Allele Sequence to capture the entire region of ambiguity. Unlike LiteralSequenceExpression normalization, however, the region of ambiguity is defined by full-length repeat subunits. The Alternate Allele Sequence is also expanded in this way, but is represented by altering the RepeatedSequenceExpression.count attribute, rather than the seq_expr attribute.
The above only applies if RepeatedSequenceExpression.seq_expr is set to a LiteralSequenceExpression object. If the RepeatedSequenceExpression.seq_expr is instead a DerivedSequenceExpression, the Allele SHOULD be returned as-is.
References
- 1
Holmes JB, Moyer E, Phan L, Maglott D, Kattman B. SPDI: Data Model for Variants and Applications at NCBI. Bioinformatics. 2019. doi:10.1093/bioinformatics/btz856
- 2
Wagner AH, Babb L, Alterovitz G, Baudis M, Brush M, Cameron DL, …, Hart RK. The GA4GH Variation Representation Specification (VRS): a Computational Framework for the Precise Representation and Federated Identification of Molecular Variation. bioRxiv. 2021. doi:10.1101/2021.01.15.426843